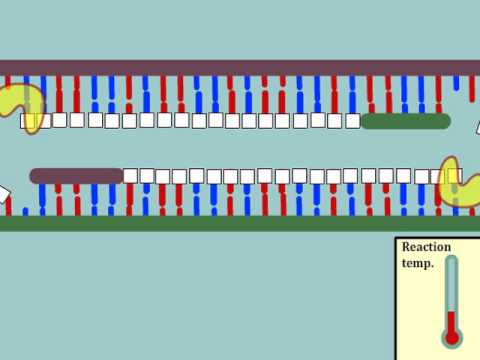
聚合酶鏈反應(PCR)介紹--多語言字幕。 (Intro to Polymerase Chain Reaction (PCR) - Multi-Lingual Captions)
Cheng-Hong Liu 發佈於 2021 年 01 月 14 日  沒有此條件下的單字
沒有此條件下的單字US /spɪˈsɪfɪk/
・
UK /spəˈsɪfɪk/
US /ˈprɛznt/
・
UK /'preznt/
- adj.出席;在場的;目前的
- n.正在進行的;現在時態;目前的;禮物
- v.t.介紹;主持;介紹;展現;贈送
- v.i.出現
- v.t.使準備好; 在...上塗底色;引爆
- adj.品質最好的;第一流的;最重要的;最合適的;質數
- n.黃金時期;鼎盛時期;底漆;質數;最優惠利率
US /riˈækʃən/
・
UK /rɪ'ækʃn/