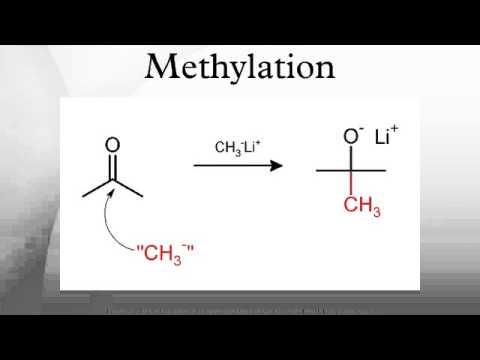
甲基化 (Methylation)
wehou 發佈於 2021 年 01 月 14 日  沒有此條件下的單字
沒有此條件下的單字US /spɪˈsɪfɪk/
・
UK /spəˈsɪfɪk/
US /ˌɪndəˈvɪdʒuəl/
・
UK /ˌɪndɪˈvɪdʒuəl/
- n. (c.)個人;單個項目;個體;個人賽
- adj.個人的;獨特的;個別的;獨特的
US /ɪkˈsprɛʃən/
・
UK /ɪk'spreʃn/
- n. (c./u.)表達;措辭;表情;表達式;表現;表現
US /ˈɛvɪdəns/
・
UK /'evɪdəns/
- n. (u.)證據;證據 (法律)
- v.t.表明;證明